Pseudomonas aeruginosa (see figure 1) is a human pathogen that colonizes the respiratory and gastrointestinal tract, where it can cause life-threatening infections in patients with a compromised immune system, such as cancer or cystic fibrosis patients. P. aeruginosa is resistant to many antibiotics. Current treatments comprise antibiotic chemotherapy and bacteriophage therapy. However, There are some setbacks to these therapies. The chemotherapy kills many kinds of bacteria, upsetting a healthy human microbiome, and the bacteriophage therapy relies on the use of a virus, which has a limited therapeutic potential as the host can develop specific antibodies against the virus.
Figure 1: Scanning electron microscope (SEM) image of P. aeruginosa.
(Source: visualphotos)
Maybe synthetic biology can come to the rescue?
A new study details the engineering of E. coli so that it can locate and destroy P.aeruginosa. In a competitive environment, the pathogen produces pyocins, self-made antimicrobials, effective against closely related species. Research has shown that one of these pyocins, known as pyocin S, is shows strong antibacterial activity against P.aeruginosa. So, engineering E. coli to produce this pyocin is a good start.
But, of course, this pyocin should only be produced in the presence of the pathogen. To ensure this, the quorum sensing mechanisms P.aeruginosa employs are exploited. Quorum sensing is a chemical signaling cascade that regulates several physiological activities, such as cell motility, virulence, and so on. The process is mediated through chemicals known as autoinducers, and their concentration depends on the pathogen density. Thus, allowing the engineered E. coli to recognize these chemicals would enable it to ‘time’ the release of the pyocins.
Here’s how the complete process works (see figure 2). The engineered E. coli contains lasR proteins that recognize the autoinducer produced by P. aeruginosa, known as3OC12 HSL. When the proteins bind this chemical, they turn into a P luxR activator, leading to the production of the pyocin S5 protein and the lysis E7 protein. The latter will, after a threshold concentration is reached, destroy the cell wall, releasing the pyocin S5 into the environment, killing the P. aeruginosa.
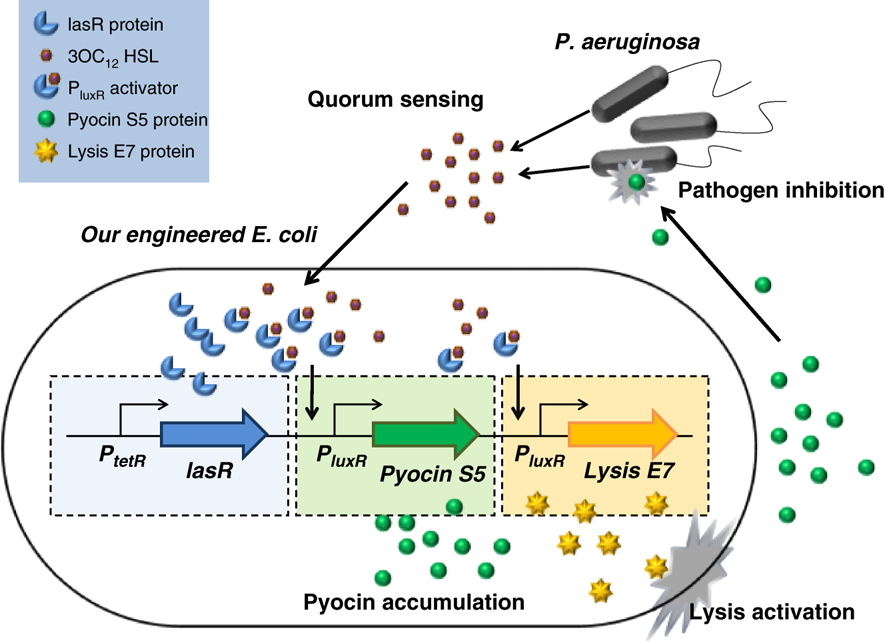
Figure 2: The 'locate and destroy' process.
(Source: Saeidi et al., 2011)
The process has been validated for a range of 3OC12 HSL concentrations, in line with those secreted by P. aeruginosa. The process has been demonstrated in both planktonic and biofilm states.
The authors conclude:
…this study presents the possibility of engineering potentially beneficial microbiota into therapeutic bioagents to arrest Pseudomonas infection. Given the stalled development of new antibiotics and the increasing emergence of multidrug-resistant pathogens, this study provides the foundational basis for a novel synthetic biology-driven antimicrobial strategy that could be extended to include other pathogens such as Vibrio cholera and Helicobacter pylori.
Reference
Saeidi, N.; Wong, C.K.; Lo, T.-M.; Nguyen, H.X.; Ling, H.; Leong, S.S.J.; Poh, C.L. and Chang, M.W. (2011). Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology. 7:521. doi:10.1038/msb.2011.55. (Click here for the article)



Comments